
Fieldwork in Sweden
Working with specimens in silico
research goals
My main research goal is to identify the mechanisms that give rise to phenotypic diversity at different levels of biological organization (i.e., within and among species). Specifically, I aim to develop a more quantitative understanding of (i) phenotypic differentiation among major groups, (ii) the origin and persistence of color polymorphisms, and (iii) phenotype-environment relationships in natural populations. To achieve these goals, I develop methodological frameworks for high-throughput phenotyping, primarily using automated image analysis (computer vision) and artificial intelligence (e.g., image encoders, vision-language models, large language models). I have applied these methods across a diverse array of biological systems (e.g., insects, crustaceans, plants, birds, etc.), integrating field surveys, controlled laboratory experiments, and natural history collections.
ongoing work
Phenomics of color evolution in butterflies
Butterfly wings provide an ideal system to study the origins of phenotypic diversity and evolutionary diversification, particularly using computer vision and AI: most phenotypic variation is along two dimensions, and therfore well suited for image-based analysis. Moreover, their modular wing patterns give rise to astonishing combinatorial complexity that is challenging to capture manually, but efficiently handled by vision encoders. Additionally, their extensive representation in natural history collections and their iconic status make them a powerful model for integrating computer vision and evolutionary biology. Using large image databases from natural history museum collections, and cutting edge foundational AI, I am empirically testing theories on the adaptive significance of wing color patterns—such as aposematism, camouflage, and mating displays.

Natural history museum collections are a rich source of organismal phenotypes.
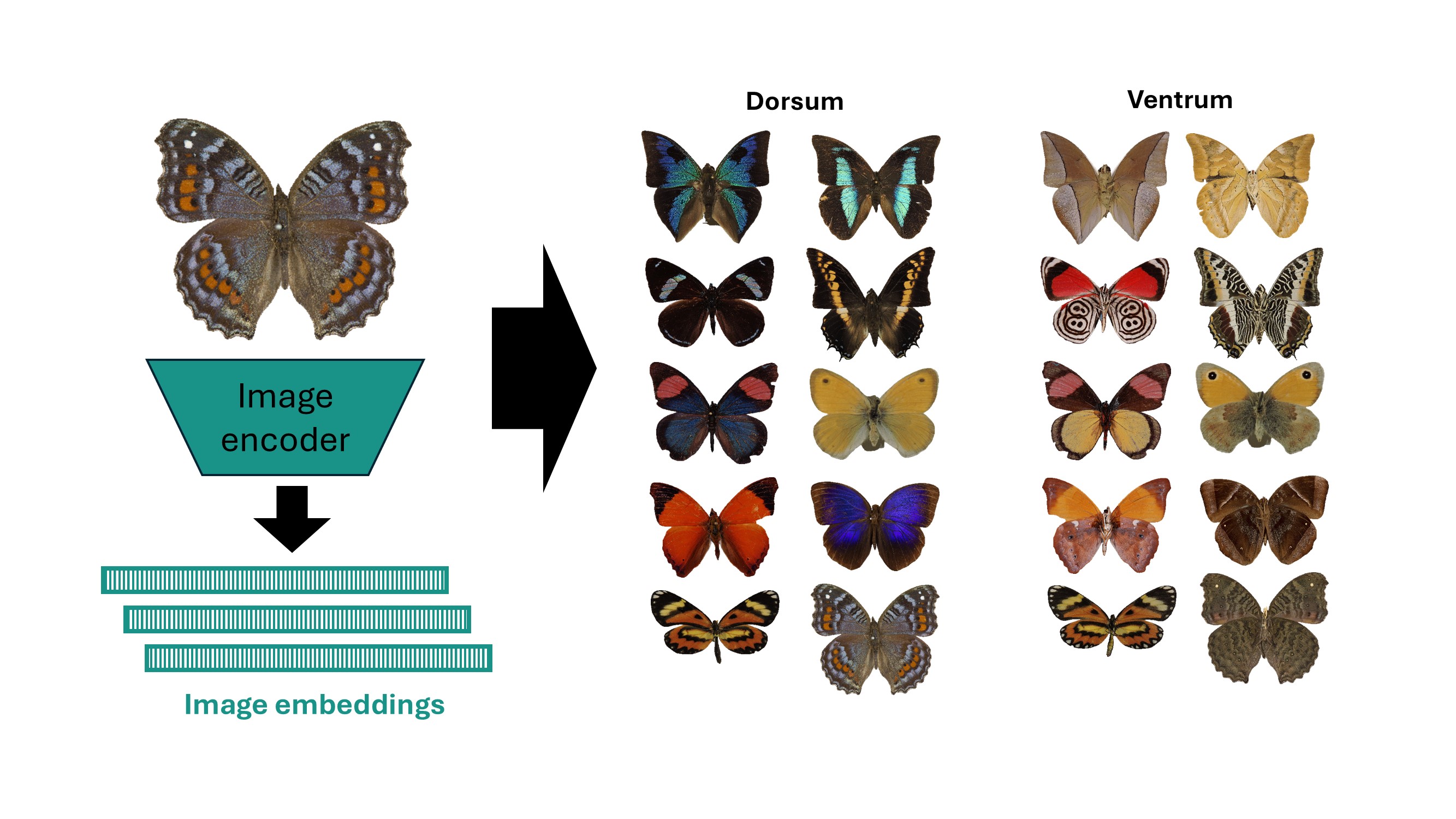
Foundational AI converts phenotypic information to generic features.
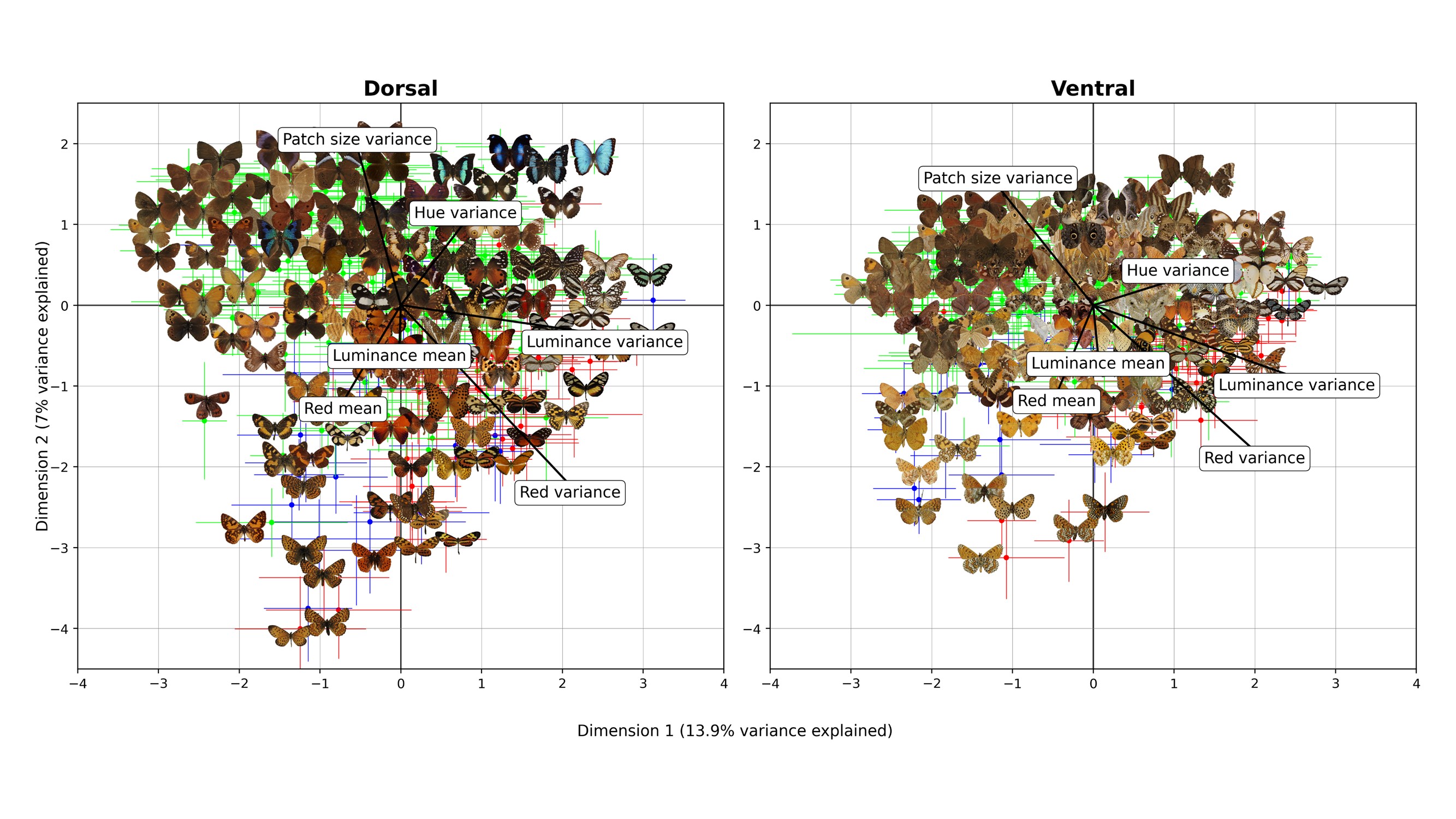
Visual feature space is mapped out with tangible metrics.
Mapping out the adaptive landscape of colour polymorphisms
Color polymorphisms are an important source of intra-specific phenotypic diversity, but the ecological and evolutionary implications of discrete vs. continuous varation within and among morphs is still poorly understood. While often rooted in discrete genetic mechanisms, the resulting phenotypic variation can be continuous, and different morphs are often overlapping along multiple phenotypic dimensions in phenotypic space. Indeed, color polymorphisms are frequently associated with variation in other traits, such as morphology and behavior. By leveraging long term image datasets and AI-based computer vision, I explore the complex phenotypic landscapes of color polymorphisms, uncovering how continuous and discrete variation interact with fitness, selection, and evolutionary trajectories.
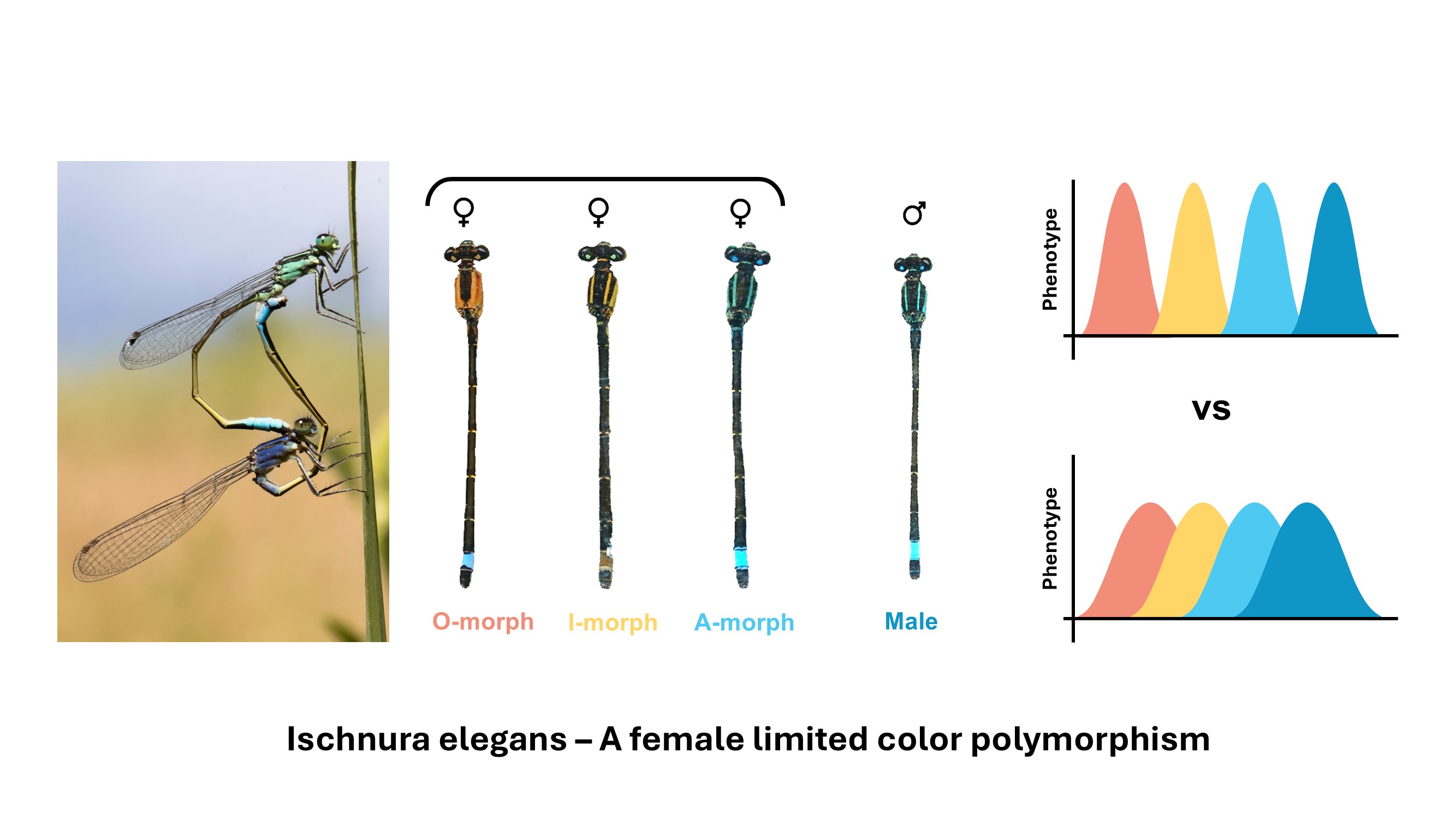
Color polymorphism in Ischnura elegans is female limited: 3 female color morphs vs 1 male morph.
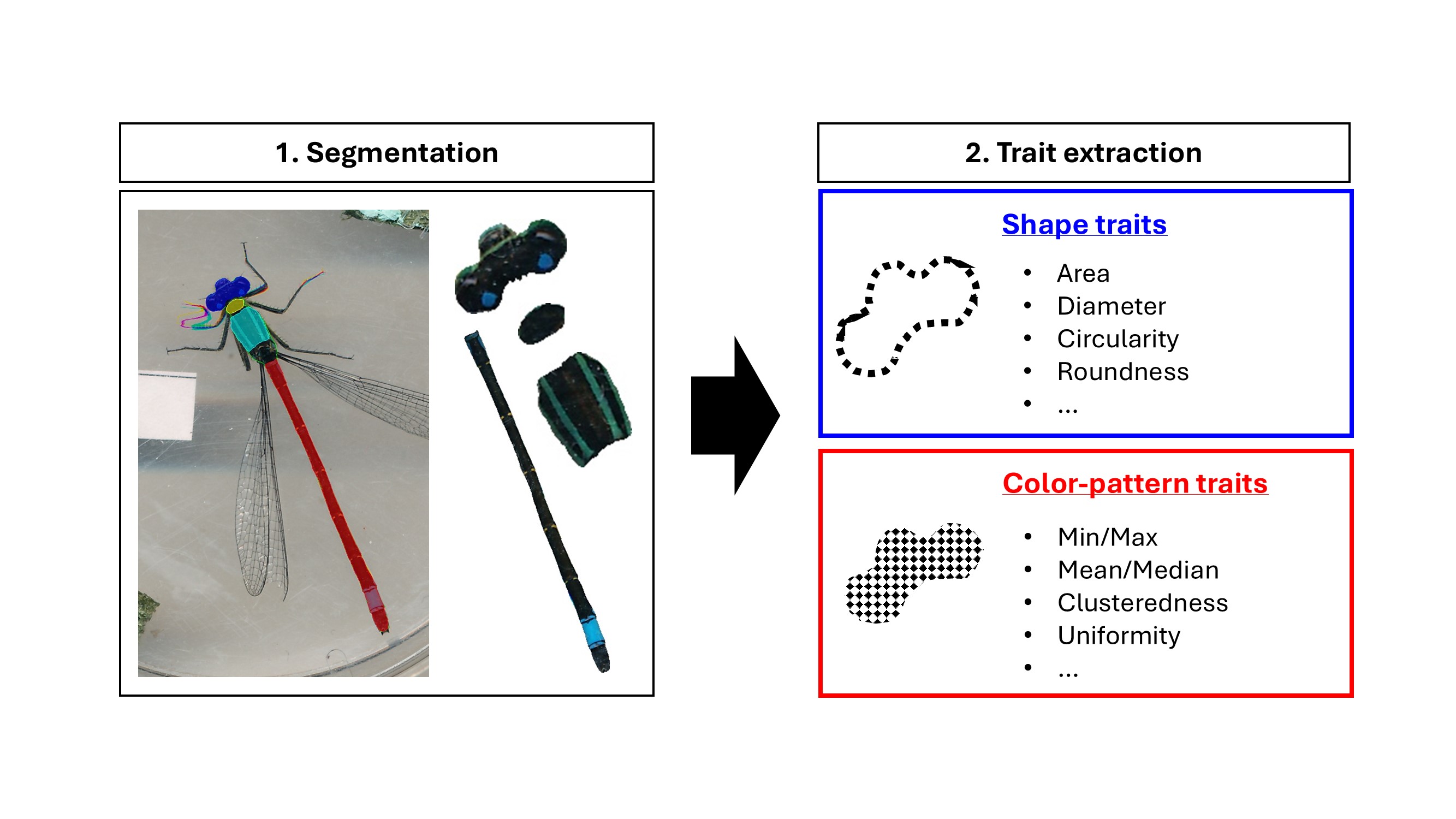
computer vision extracts aspects of shape and coloration in high throughput.
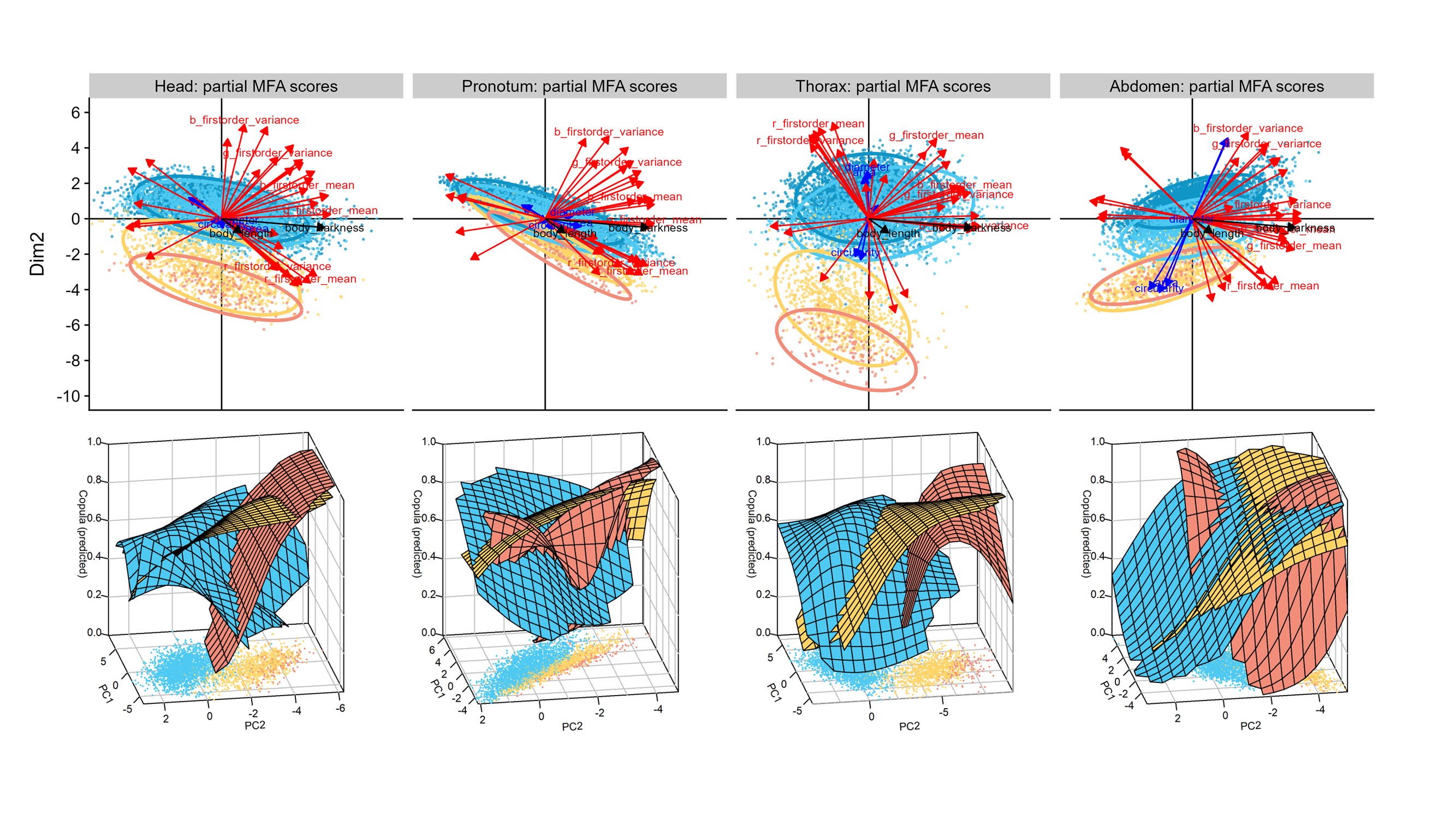
The combination of phenotypic and mating information can detect singals of sexual selection.
The phenomicist's toolkit: developing modern tools for phenotyping
At large, my research program aims at a stronger integration of phenotypic information in ecological and evolutionary resarch. To do so, I develop high-throughput phenotyping methods that leverage computer vision to extract rich, high-dimensional phenotypic data from images. These pipelines focus on userfriendliness to enable efficient and reproducible trait quantification across diverse biological systems (e.g., phenopype), and provide access to state-of-the-art AI techniques (e.g., BioEncoder). By building and applying these tools, I aim to overcome bottlenecks in phenomic data collection and make large-scale phenotypic data acquisition accessible to users from the domain of biology, ecology and evolution, with little to no computational background.
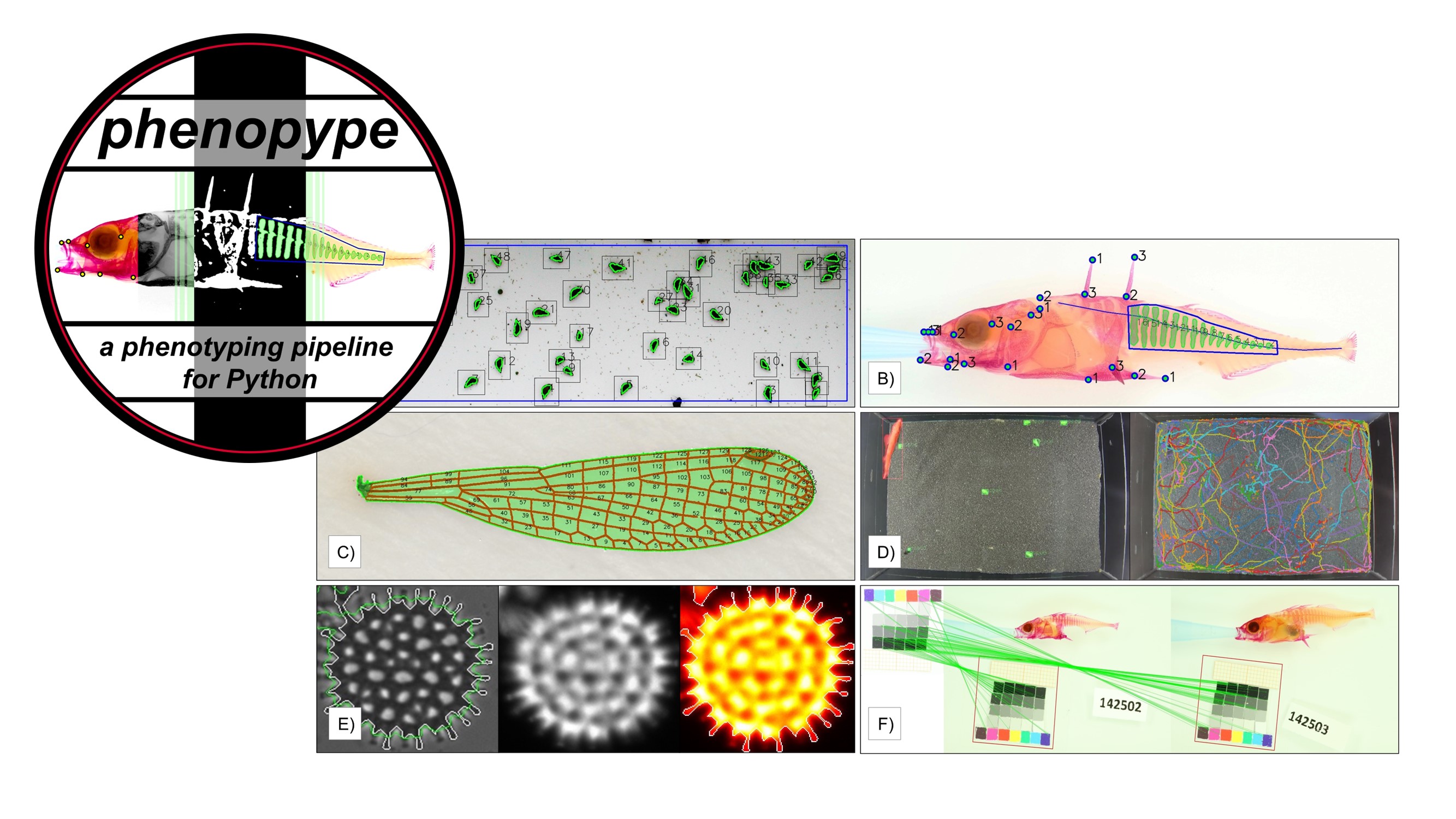
phenopype: A phenotyping pipeline for Python
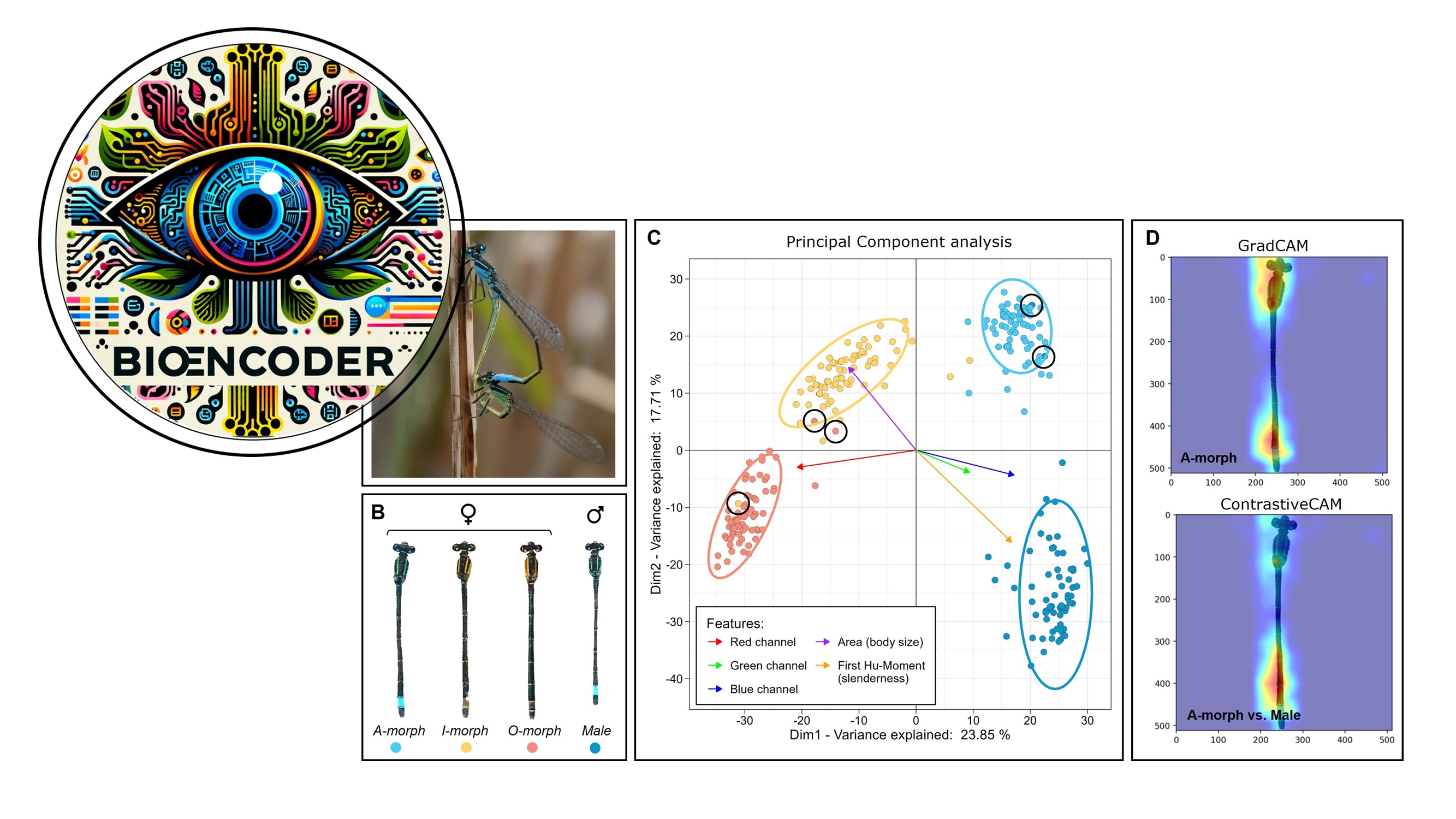
BioEncoder: A metric learning toolkit for comparative organismal biology
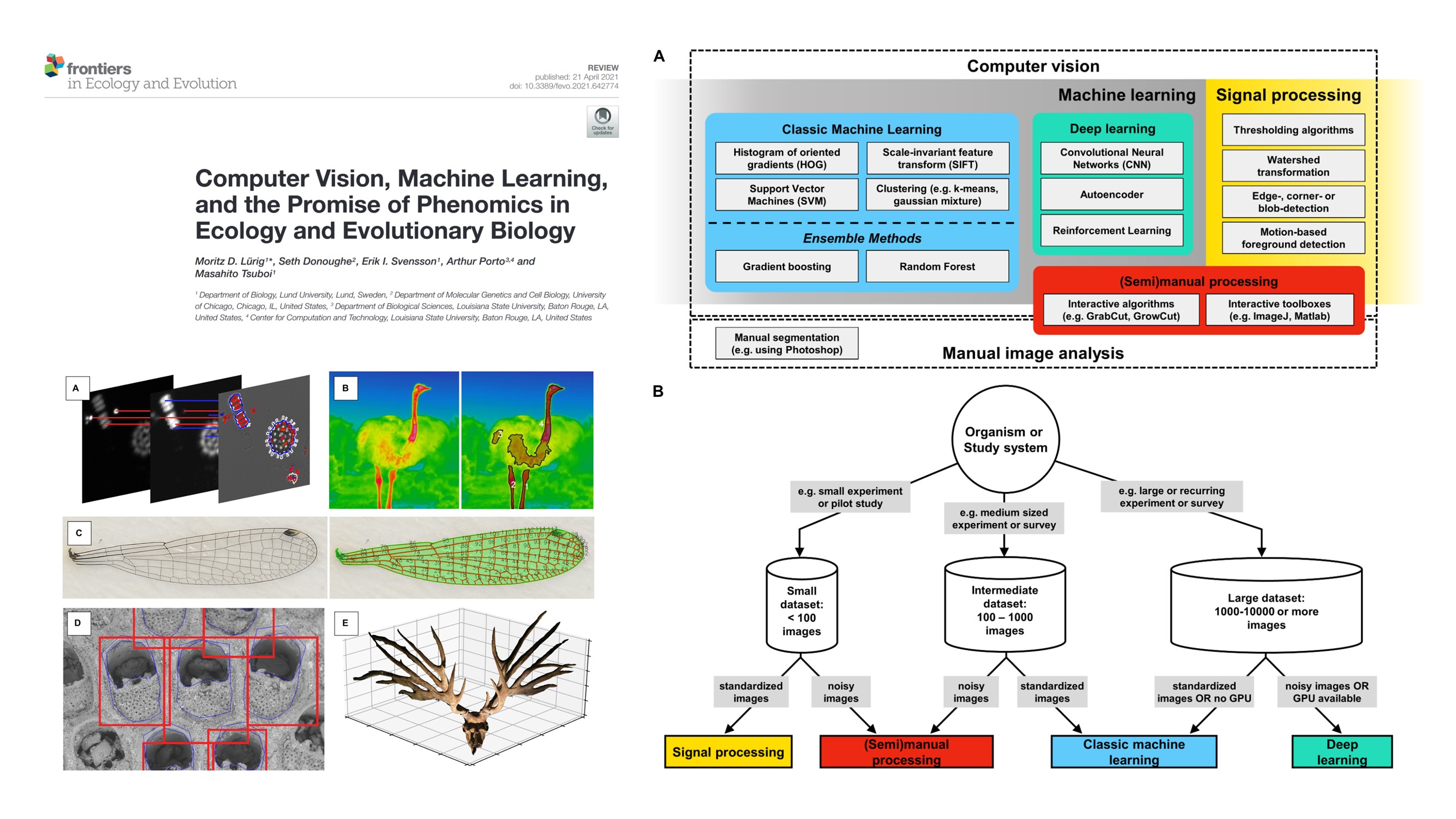
Review on the use of computer vision for phenomics in ecology and evolutionary biology
- Lürig, M.D., Di Martino, E. and Porto, A. (2024) ‘BioEncoder: A metric learning toolkit for comparative organismal biology’, Ecology letters, 27(8), p. e14495.
 [DOI]
[DOI] - Lürig, M.D. (2022). phenopype: A phenotyping pipeline for Python. Methods in Ecology and Evolution, 13(3), 569–576.
 [DOI] [PDF]
[DOI] [PDF] - Lürig, M.D., Donoughe, S., Svensson, E.I., Porto, A., and Tsuboi, M. (2021). Computer Vision, Machine Learning, and the Promise of Phenomics in Ecology and Evolutionary Biology. Frontiers in Ecology and Evolution 9:642774.
 [DOI] [PDF]
[DOI] [PDF]
previous work
The adaptive landscape of pigmentation and nutrition in freshwater isopods
The pigmentation of the freshwater isopod Asellus aquaticus is thought to be shaped visual predation along a gradient of background darkness. In a mesocosm experiment where me manipulated background darkness and predation pressure, we found that fish predation reduced isopod density, but pigmentation was primarily influenced by macrophyte presence, suggesting an environmental rather than predator-driven effect. A subsequent laboratory experiment revealed that pigmentation responds to dietary protein levels, with higher protein intake accelerating pigmentation development but increasing juvenile mortality under rapid growth. These findings indicate that dietary-driven plasticity can shape phenotypic variation and survival outcomes, even in the absence of predators. Together, these results highlight the complex interplay between genetic background, environmental conditions, and life-history traits in shaping isopod pigmentation.
Asellus aquaticus is becoming a versatile study system to investigate long-standing questions at the interface of ecology, evolution, and development. Findings from this and other work on the relevance of isopod pigmentation and development are summarized in a review article on A. aquaticus (Lafuente et al. 2021).

Variation in habitat background darkness in Lake Lucerne at Eawag Kastanienbaum

Variation in pigmentation in freshwater isopods is thought to be an adaptive response to predation along a gradient of background darkness.

Mesocosm experiment to manipulate background darkness and predation pressure.

Imaging setup for computer-vision-based quantification of pigmentation in isopods

Differences in pigmentation between high (top) and low protein (bottom) supplement during ontogeny.
- Lürig, M.D., Narwani, A., Penson, H., Wehrli, B., Spaak, P., and Matthews, B. (2021). Non-additive effects of foundation species determine the response of aquatic ecosystems to nutrient perturbation. Ecology 102(7), e03371. [DOI] [PDF]
- Lürig, M.D., Best, R.J., Dakos, V., and Matthews, B. (2020). Submerged macrophytes affect the temporal variability of aquatic ecosystems. Freshwater Biology 66(3), 421-435. [DOI] [PDF]
- Lafuente, E., Lürig, M.D., Rövekamp, M., Matthews, B., Buser, C., Vorburger, C., & Räsänen, K. (2021). Building on 150 Years of Knowledge: The Freshwater Isopod Asellus aquaticus as an Integrative Eco-Evolutionary Model System. Frontiers in Ecology and Evolution, 9.
 [DOI] [PDF]
[DOI] [PDF]
Foundation Species and the Stability of Aquatic Ecosystems
Foundation species shape ecosystem structure and function, but their effects can vary across timescales and in response to environmental change. Using a series of outdoor mesocosm experiments and high-frequency multi-parameter sensor technology we found that macrophytes reduce phytoplankton biomass, increase DOM accumulation, and influence ecosystem variability over time. However, when co-occurring with mussels, their effects on nutrient dynamics became less predictable, leading to a counterintuitive increase in phytoplankton biomass. These results show that while foundation species can stabilize ecosystems, their interactions may generate unexpected nonlinear responses to environmental perturbations.

High frequency multi-parameter sondes (WTW/YSI) during calibration.

Outdoor experimental pond ecosystems (20x 15000L) at Eawag Dübendorf.
- Lürig, M.D., Narwani, A., Penson, H., Wehrli, B., Spaak, P., and Matthews, B. (2021). Non-additive effects of foundation species determine the response of aquatic ecosystems to nutrient perturbation. Ecology 102(7), e03371. [DOI] [PDF]
- Lürig, M.D., Best, R.J., Dakos, V., and Matthews, B. (2020). Submerged macrophytes affect the temporal variability of aquatic ecosystems. Freshwater Biology 66(3), 421-435. [DOI] [PDF]
 [DOI]
[DOI]  [DOI] [PDF]
[DOI] [PDF]  [DOI] [PDF]
[DOI] [PDF]  [DOI] [PDF]
[DOI] [PDF] 
















